GlobDB release 226 methods
Dereplication of the underlying databases
11 of the 14 data sources for the GlobDB release 226 (GTDB, mOTU, RBG, SPIRE, GEM, MGnify, GOMC, SMAG, MRGM/HRGM2, cFMD, SHGO, AMXMAG, GFS) provide a dereplicated set of genomes, using a 95 % average nucleotide identity (ANI) dereplication criterion. Since the underlying data for each database is distinct, there may be centroids between datasets that belong to the same species by the operational definition of 95 % ANi similarity, but between them represent a cluster of genomes that extends beyond that operational species boundary. Thus it is possible that the genomes that two centroids represent wouldn't all be classified to the same species if all data was processed together.
The GlobDB therefore conservatively dereplicates the underlying (already dereplicated) datasets at 96 % ANI over 50 % aligned fractions (AF). We have chosen these cutoffs after inspection of ANI histograms and crossplots of ANI vs AF values (see examples below), showing a clear ANI gap between the dereplicated datasets starting at approximately 96% ANI.
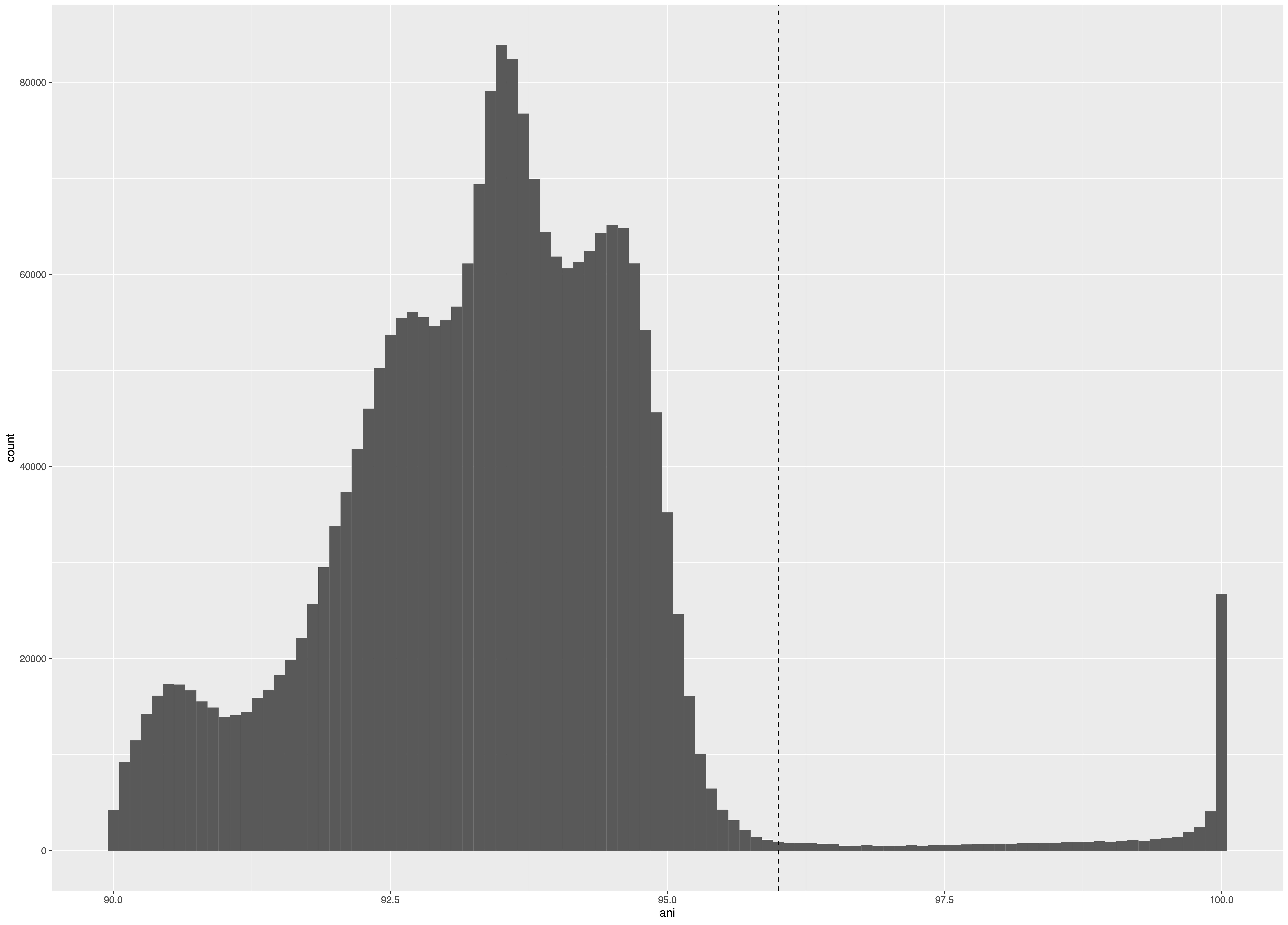
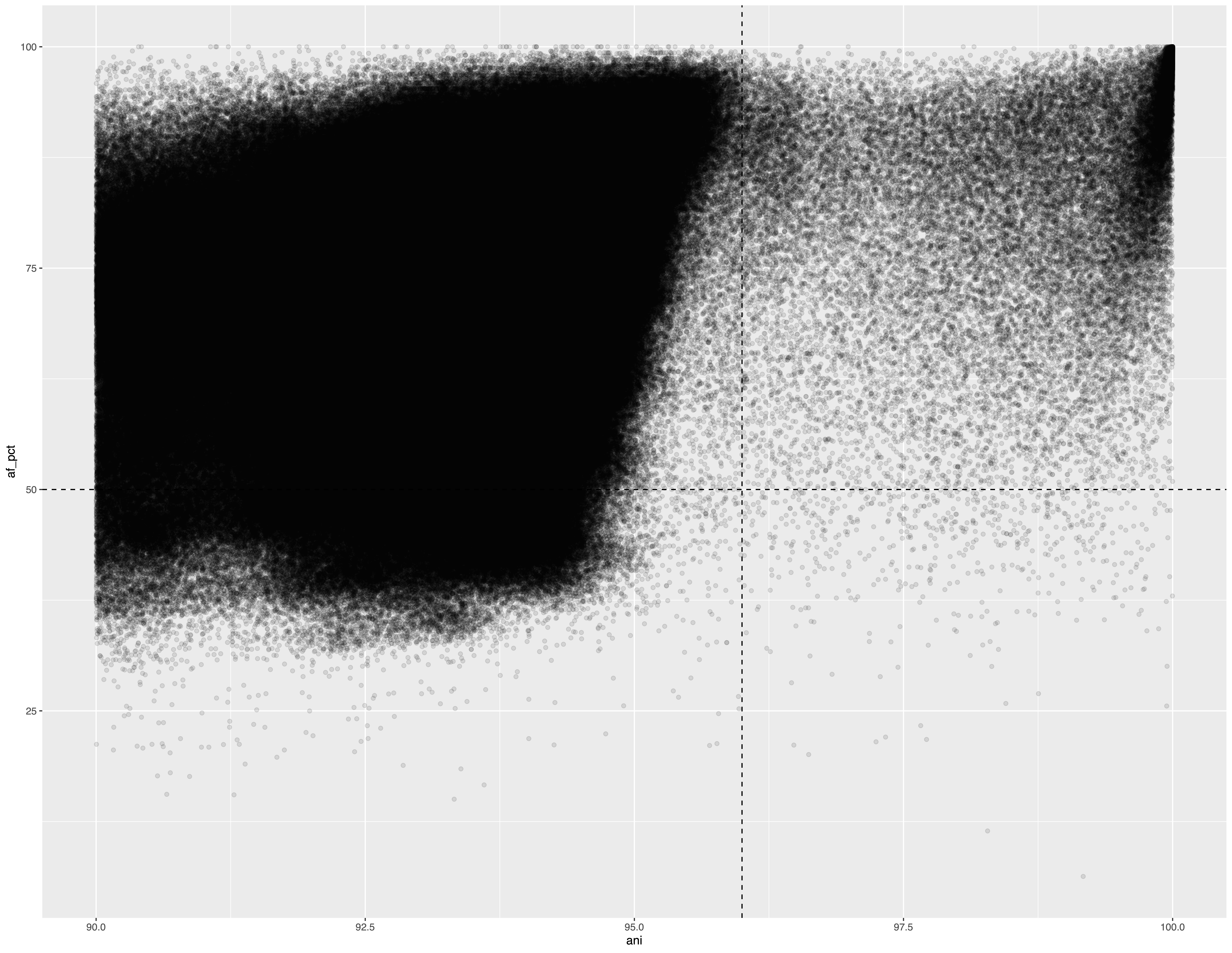
Left: Histogram showing the pairwise ANI values of the GTDB (release 226) and the mOTU 4.0 species representatives. Right: crossplot of ANI and aligned fractions (AF) of the GTDB (release 226) and the mOTU 4.0 species representatives. Dashed lines indicate the cutoffs for inclusion in the GlobDB at 96% ANI and 50% AF.
Furthermore, the order of priority for inclusion in the GlobDB is as follows:
- All GTDB species representatives are included.
- The other datasets are sequentially dereplicated. This means that the MAGs of the second dataset that are not represented in the GTDB are added to the GlobDB, then the MAGs from the third dataset that are not represented in this new combined set are added to the GlobDB, and so on.
- The order of sequential dereplication in release 226 is GTDB, mOTU, RBG, SPIRE, GEM, MGnify, GOMC, SMAG, MRGM/HRGM2, cFMD, SHGO, AMXMAG, GFS
No species dereplicated set was available for the cFMD, SHGO, and AMXMAG datasets. To account for this, the full dataset was first dereplicated against the GlobDB, followed by dereplication of the remainder of the genomes (>96% ANI and >50% AF) using dRep (v3.4.2).
Dereplication of the data sources is done using fastANI, version 1.3.4, run with the 3000bp fragment length default.
The complete final dataset was then assessed for quality using CheckM2 and anvi'o, which removed 420 genomes from the dataset. There were four reasons for exclusion of genomes at this stage, in this order, with numbers in brackets:
1) genomes that failed the annotation process (7)
2) genomes with a CheckM2 completeness under 40 % (341)
3) genomes with an anvi'o domain designation EUKARYA (68)
4) genomes with a total length over 20M bp (4)
For convenience in later use, the names of the genomes in the all datasets except the GTDB, MGnify, and MRGM/HRGM2 are standardized to GlobDB identifiers that represent their origin and are propagated throughout the GlobDB resources. Dictionary files to relate the GlobDB IDs back to the identifiers from the original publications are available in the Downloads page.
Generation of anvi'o databases and annotation
After dereplication, the resulting genome fasta files are turned into anvi'o contigs databases and basic annotation is performed.
First, contig names are standardized using anvi'o with anvi-script-reformat-fasta and the options: --seq-type NT # specifies the sequence type--simplify-names # standardizes the contig name format--overwrite-input # overwrites the source file--prefix <GlobDB_ID> # prepends the GlobDB ID to the contig name
After this, contigs databases are created using anvi-gen-contigs-database, using the GlobDB ID as database name (-n flag). Basic annotations on these contigs databases are run using HMMs integrated with anvi'o. Information on the sources of HMMs that are included in anvi'o can be found here. Addditionally, KEGG, COG, Pfam and CAZy annotations are performed. Please see the links in the file descriptions on the Downloads page for more information on the anvi'o implementation of these annotations. One specific thing to note is that for KEGG annotations, anvi'o employs a heuristic to add (likely) true positive hits that were missed during the first pass, described in more detail here.
These contigs databases are then populated with annotations using the following commands:anvi-run-hmms # annotates rRNA genes, marker genes, and tRNAs anvi-run-kegg-kofams # annotates coding regions with kegg kofamsanvi-run-pfams # annotates coding regions with pfamsanvi-run-cazymes # annotates coding regions with dbCAN2 CAzymesanvi-run-ncbi-cogs # annotates coding regions with COGs (2020 update)
Next, protein fasta files and gff files for KEGG, COG and Pfam annotations are exported from the contigs databases
These files can also be generated from the anni'o databases directly using the anvi-get-sequences-for-gene-calls command with different flags.
# protein fasta--get-aa-sequences --wrap 0 --defline-format {contigs_db_project_name}___{gene_caller_id} # gff with COG annotation--export-gff3 --annotation-source COG20_FUNCTION
Clustering amino acid sequences and generating pLM embeddings
The protein fastA files of all GlobDB genomes were concatenated into a single fastA file for clustering using Linclust as integrated in MMseqs2 (Release 17-b804f). Clustering was done at 40 % sequence identity (--min-seq-id 0.4) over 80 % of the length of both query and target sequence (--cov-mode 0 -c 0.8).
Representatives of clusters consisting of at least two proteins were then used for protein language model (pLM) embedding using the ProtT5-XL-U50 protein large language model. A more detailed description of the embedding process, as well as the scripts used to generate the embeddings are available in this github repository.
Establishing the GlobDB Taxonomy
De novo genome trees for bacterial and archaeal genomes from novel species were generated using GTDB-tk v2.4.0 de_novo_wf (Chaumeil et al. 2022). Novel clades were identified through a custom algorithm based on relative evolutionary divergence (RED), similar to that described in Supplementary Figure S1 from Chaumeil et al. 2020. Firstly, RED values were calculated by PhyloRank v0.1.12 using the de novo genome trees from GTDB-tk. Next, the RED values and genome trees were processed using a custom script (see name_clades.py here). Genomes were sequentially assigned to either pre-existing or novel clades, ordered by genome quality. For each genome, this involved traveling up the tree until a node was reached within the RED-bounds of a new taxa level (e.g. phylum, class, order, etc.) for which the genome has not yet been assigned. Parents and children of the node were checked to find if any were closer to the median RED of that taxa level (from GTDB R226) than the current node. If the current node was the closest node, it is annotated as a new clade, with the current genome being assigned as the genome representative. This continued until a node was reached that had an offspring from GTDB (or until reaching a previously labelled node). The remaining taxonomy was assigned based on the median taxonomy of the GTDB offspring from that node. A snakemake workflow implementing the above can be found in this github repository.
Matching genome IDs to culture availability
The BacDive python API was queried to linked species representatives in the GlobDB to their respective isolates deposited in major culture collections. A search of BacDive was performed for genomes whose accession numbers began with the prefix "GC" and the entries associated with these genomes retrieved. These isolates, and their culture collection numbers, were then matched to the representatives in the GlobDB based on the genome accession. This match was performed on the numeric part of the genome accession, which remains invariant between GenBank and RefSeq. This ensured that isolates submitted to culture collections and originally linked to a GenBank accession could still be correctly associated, even if the genome was later upgraded to a RefSeq accession not reflected in BacDive.