Welcome to the GlobDB genomes database
This website hosts the GlobDB, a dereplicated set of species representative microbial genomes. The genomic era offers great opportunities for microbial genome analyses, and individual (meta)genome studies can generate thousands of microbial genomes. Although the International Nucleotide Sequence Database Collaboration (INSDC) databases are available to store these datasets, depositing assembled and binned metagenome assembled genomes (MAGs) to INSDC databases doesn't always happen. The GlobDB aims to integrate 14 genome resources that are currently not (yet) consolidated otherwise.
Our paper describing the GlobDB has now been published in Bioinformatics Advances.
GlobDB features
The 14 genome resources (see below) predominantly provide "species dereplicated" genome catalogs. These catalogs are then further sequentially dereplicated (in the order they are listed below) and the dereplicated set is processed in a standardised way to yield a comprehensive dataset that can be used for further analyses. Currently, the GlobDB comprises 306,260 (partial) microbial genomes after dereplication of the 14 source datasets.
For all 306,260 genomes, the GlobDB features:
- Genome fasta files with standardized names and IDs
- Anvi'o contigs databases, annotated with KEGG/COG/Pfam/dbCAN2
- Amino acid fasta files with standardized identifiers
- GFF files for coordinates of genes
- A full 7-level taxonomy built upon and extending the GTDB taxonomy
- Basic genome statistics and completeness/contamination
- Whether the genome accession is linked to an isolate deposited in a major culture collection
Furthermore, the GlobDB includes:
- a SingleM metapackage linked to the GlobDB taxonomy, for taxonomic profiling of metagenome datasets
- a Sylph database linked to the GlobDB taxonomy, for taxonomic profiling of metagenome datasets.
- 82,972,511 cluster representatives of proteins detected at least twice
- ProtT5 protein language model embeddings for the 82,972,511 cluster representatives
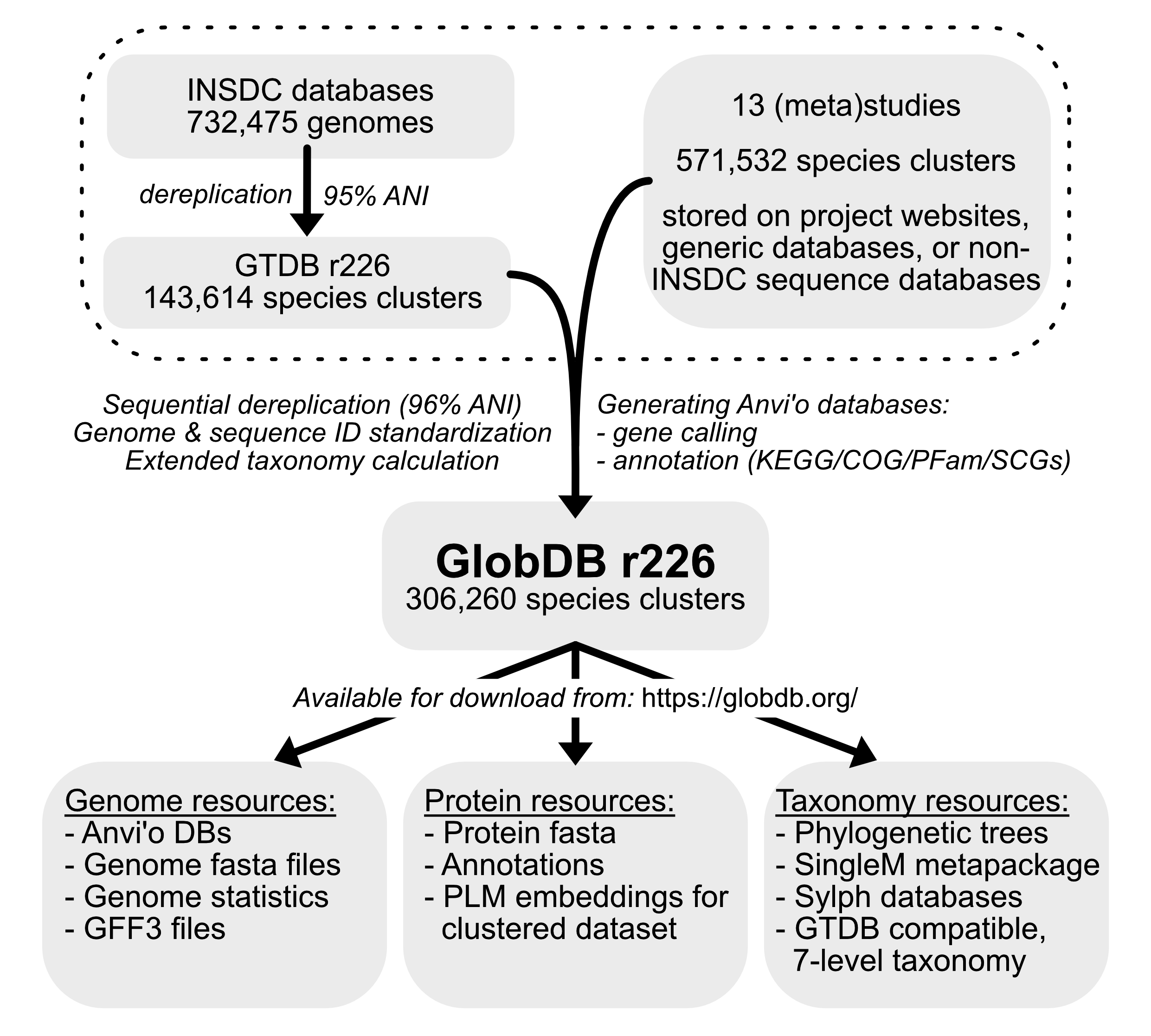
See the Methods for details on data processing, and Downloads for a description of available files.
If you use the GlobDB, make sure to cite the underlying data sources and methods as appropriate, see How to cite.
GlobDB source datasets
As of version 226, the GlobDB includes 14 data sources:
1) the species reps of the genome taxonomy database (GTDB), sourced from NCBI genome.
2) the species reps of the mOTUs 4.0 database (mOTU)
3) the species reps of the searchable, planetary-scale microbiome resource (SPIRE).
4) the species reps of the Bin Chicken Rare biosphere genomes collection (BCRBG)
5) the species reps of the genomic catalog of earth's microbiomes (GEM).
6) the species reps of the 13 MGnify biome MAG catalogs
7) the species reps of the global ocean microbiome genome catalogue (GOMC)
8) the species reps of the genomic catalog of soil microbiomes (SMAG).
9) the species reps of the Tibetan Plateau microbial catalog (TPMC)
10) the species reps of the Mouse and Human Reference Gut Microbiome (MRGM & HRGM2) catalogs
11) the MAGs of the curated Food Metagenomic Data (cFMD) resource
12) the MAGs of the sheep and goat gut microbiome compendium
13) the MAGs of the genome catalog of anammox microbiotas
14) the species representatives of the Glacier fed streams (GFS) genome catalog
Updates, versions, maintenance
The current GlobDB version is 226. The GlobDB follows the GTDB update schedule, which is currently once per year. The version numbering is linked to GTDB, which is in turn taken from to the NCBI RefSeq versioning.
The GlobDB is maintained by Daan Speth, senior scientist at the centre for microbiology and environmental systems science (CeMESS) at the university of Vienna, with contributions from colleagues at CeMESS and the centre for microbiome research at the Queensland University of Technology (QUT). See contributors for more information.
The GlobDB is hosted on the life science compute cluster (LiSC) of the University of Vienna. For any questions related to the GlobDB, please contact Daan Speth
License
the GlobDB propagates the licenses of the underlying data sources, and is licensed under CC BY-SA 4.0.
This means you are free to:
Share — copy and redistribute the material in any medium or format for any purpose, even commercially.
Adapt — remix, transform, and build upon the material for any purpose, even commercially.
Under the following terms:
Attribution — You must give appropriate credit , provide a link to the license, and indicate if changes were made . You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original.
See https://creativecommons.org/licenses/by-sa/4.0/ for full license details